AI-Based Rheumatic Heart Disease Diagnosis
Background
Rheumatic Heart Disease (RHD) remains a leading cause of cardiovascular morbidity in low-resource settings. Our work at DSAIL focuses on building deployable AI systems that support early diagnosis of RHD using echocardiographic data—bridging clinical needs with scalable machine learning.
Data Annotation and View Classification
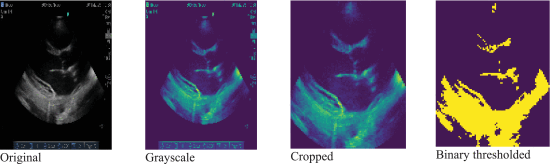
Our initial work tackled the challenge of identifying the parasternal long axis (PLAX) view, which is
critical for RHD screening. We developed a binary classifier using logistic regression and later CNNs to
distinguish PLAX from non-PLAX frames. To support scalable annotation, we built a web-based tool (Echo
Label) deployed on Google Cloud Platform, enabling cardiologists to tag echo frames with WHF-guided
metadata.
Highlights
- PLAX vs. non-PLAX classifier with >99% accuracy
- Echo Label app with WHF 2012 criteria integration
- GCP-based deployment for multi-user annotation
Unsupervised View Discovery
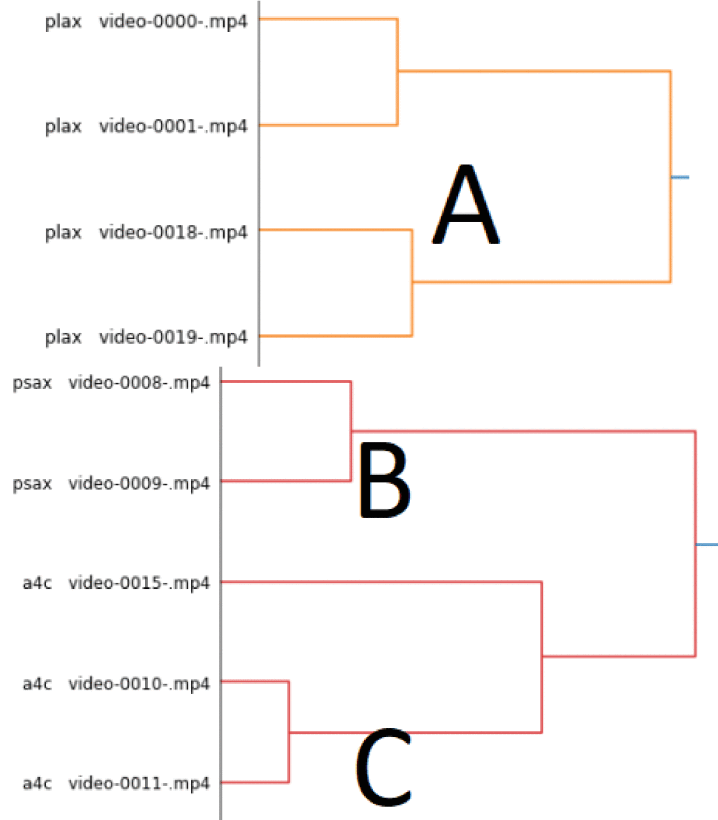
To reduce expert annotation burden, we explored unsupervised clustering of echo videos using PCA and
agglomerative hierarchical clustering. These methods grouped videos by view (PLAX, PSAX, A4C) without
labels, revealing strong potential for semi-automated annotation workflows.
Technical Methods:
- PCA for dimensionality reduction
- Ward’s linkage clustering on Euclidean distances
- Sensitivity analysis across sonography machines
Self-Supervised Multi-Task Learning
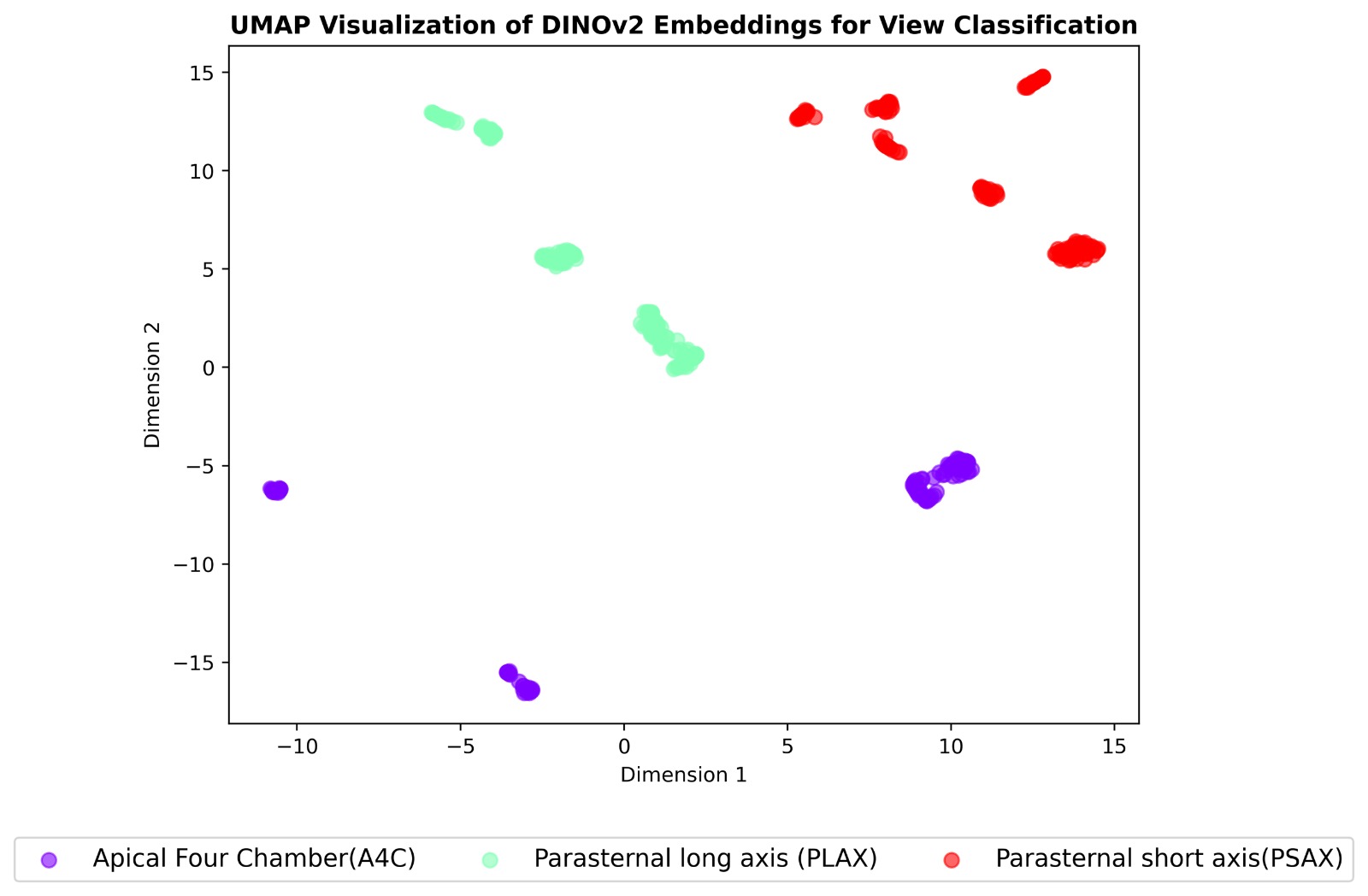
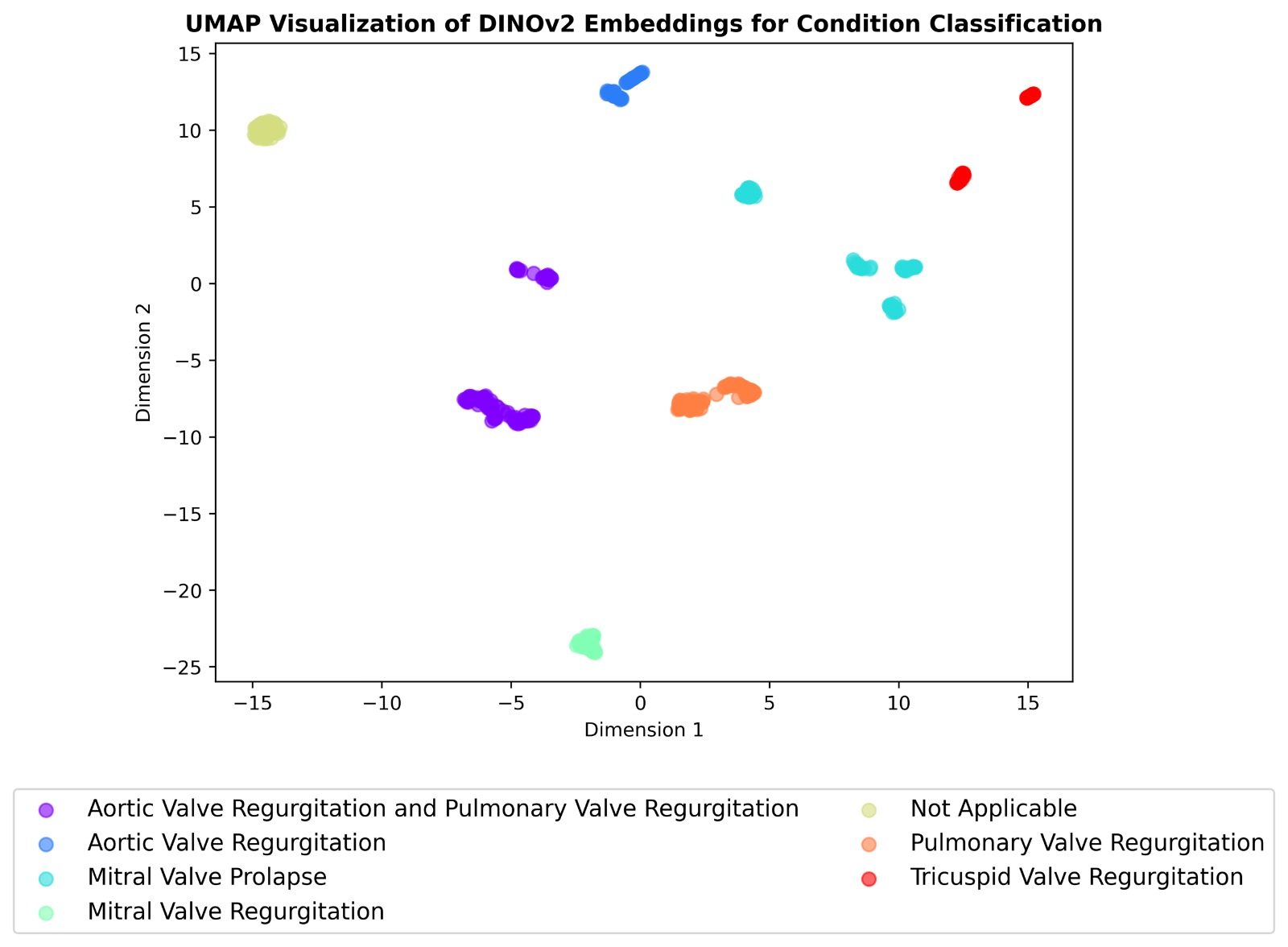
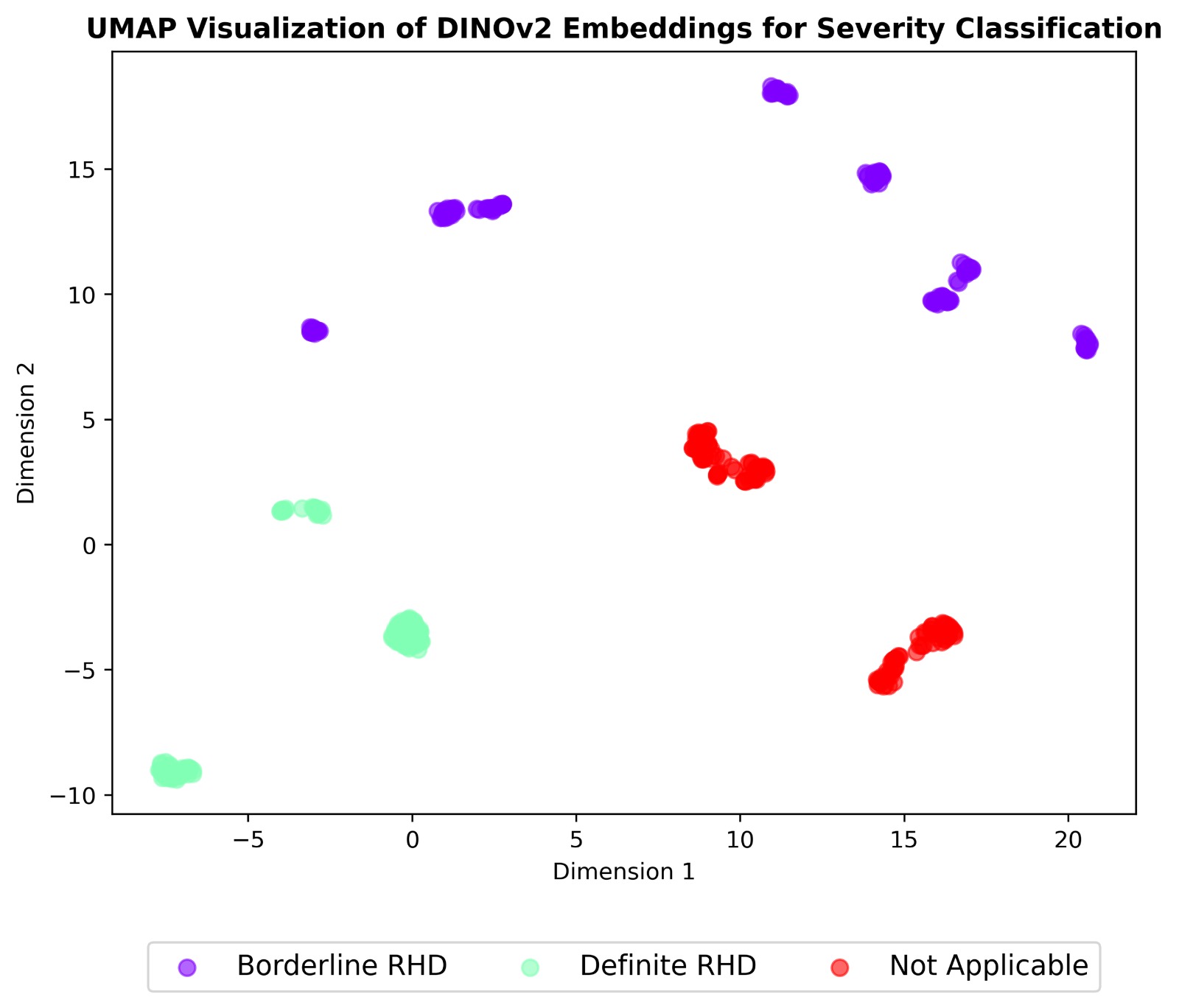
We extended our pipeline to multi-task classification of echocardiographic views, RHD conditions, and
severity using self-supervised learning. Two models were compared:
Key Components:
- SimCLR (ResNet-based contrastive learning)
- DINOv2 (Vision Transformer with self-distillation)
Both were pre-trained on 38,000+ unlabeled frames and fine-tuned on a curated set of 2,655 labeled
images. DINOv2 achieved up to 99% accuracy on severity classification, outperforming SimCLR on most
tasks.
Tasks:
- View classification (PLAX, PSAX, A4C)
- Condition detection (e.g., mitral regurgitation)
- Severity assessment (WHF 2012: normal, borderline, definite)
Next Steps
Fine-Tuning MedGemma for Clinical Deployment
We are now fine-tuning MedGemma, a multimodal vision-language model, for RHD detection in echocardiographic images. This involves adapting MedGemma to:
- Classify valvular pathology (e.g., mitral valve regurgitation)
- Interpret morphological features per WHF 2023 guidelines
- Enable on-device inference for real-time screening in clinics
Technical Direction:
- Prompt engineering for echo-specific tasks
- Evaluation on frame-level and video-level inputs
- Benchmarking against CNN and transformer baselines
Our long-term goal is to deliver interpretable, deployable AI tools that support frontline clinicians in diagnosing RHD early and accurately—especially in underserved regions. This project exemplifies how multimodal learning, clinical collaboration, and thoughtful deployment can be leveraged to address global health challenges.